Unravelling the mutational landscape in secondary glioblastoma
Secondary glioblastoma (sGBM) is a type of aggressive high-grade brain cancer that progressed from an earlier lower-grade gliomas. A comprehensive analysis of the genomic alterations in sGBM, as well as the alterations that are related to disease progression, has not been conducted due to limited data availability, hindering the discovery of druggable targets and development of precision medicine in sGBM.
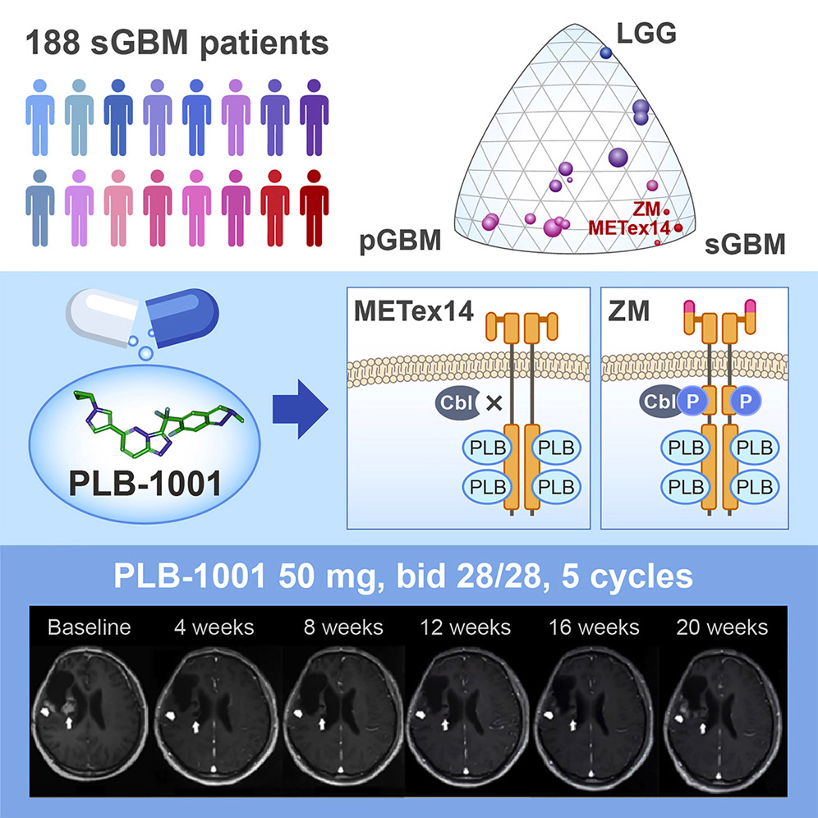
By integrating sequencing data from Beijing Tiantan Hospital and published datasets, we revealed the mutational landscape of 188 sGBM patients and identified enrichment of MET gene alterations, including MET-exon-14-skipping (METex14), PTPRZ1-MET fusions and MET amplifications, in contrast to other glioma types. In particular, METex14 is highly recurrent in sGBM (14%), and is associated with disease progression and worse prognosis. Mechanistically, METex14 alterations lead to the elimination of negative feedback regulation on MET signaling and thus result in prolonged MET activity. A MET-specific kinase inhibitor, PLB-1001, demonstrated safety and efficacy in a Phase I clinical trial, and achieved partial response in at least two chemo-resistant sGBM patients harboring MET alterations. For more details please read the published paper in Cell.
MetaNet: a machine learning method in organotropic metastases to stratify
progression risk of primary tumors
Metastasis leads to most cancer deaths, but its molecular mechanism and spatiotemporal behavior remain elusive. In this study, we asked whether genome features collected at diagnosis could be used to predict cancer development and metastasis in late stages. Accordingly, we developed MetaNet, a computational framework that integrates clinical and sequencing data from 32,176 primary and metastatic cancer cases, to assess metastatic risks of primary tumors. MetaNet achieved high accuracy in distinguishing the metastasis from the primary in breast and prostate cancers. From the prediction, we identified Metastasis-Featuring Primary (MFP) tumors, a subset of primary tumors with genomic features enriched in metastasis and demonstrated their higher metastatic risk and shorter disease-free survival. In addition, we identified genomic alterations associated with organ-specific metastases and employed them to stratify patients into various risk groups with propensities toward different metastatic organs. Remarkably, this organotropic stratification method achieved better prognostic value than the standard histological grading system in prostate cancer, especially the identification of Bone-MFP and Liver-MFP subtypes, with organotropic insights to inform organ-specific examinations in follow-ups.
Brain Arteriovenous Malformation (bAVM)
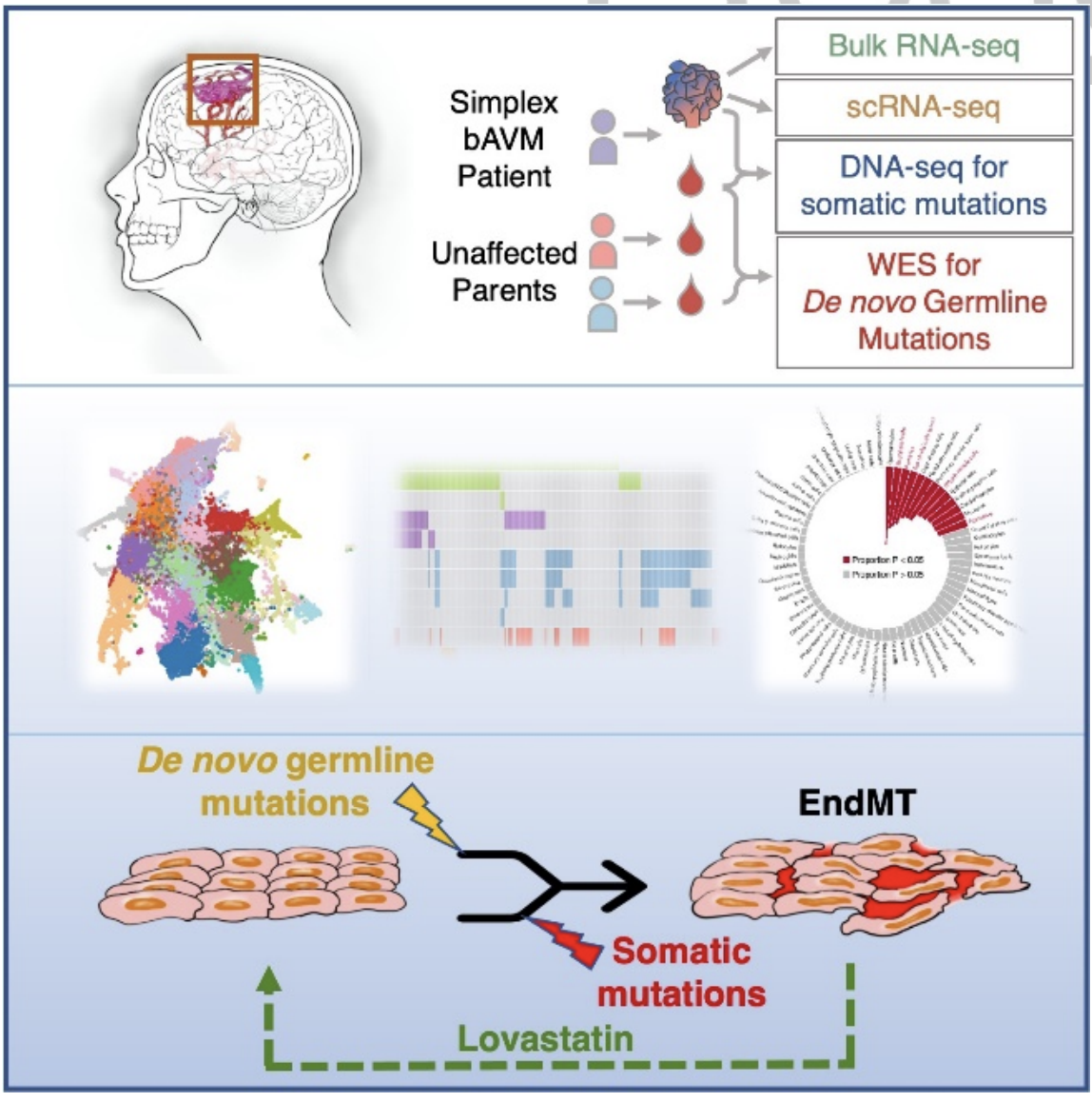
In this project, we uncovered the de novo germline and somatic mutational landscape of bAVM. Specifically, we established a comprehensive bAVM dataset from 269 patients, by performing single-cell sequencing of 17 bAVM lesions, whole-exome sequencing of germline DNA from 60 case-unaffected-parental trios, and genomic/transcriptomic sequencing of 231 bAVM lesions. For data analytics, we developed the LEMONADE algorithm (https://github.com/WangLabHKUST/LEMONADE) for ultra-deep sequencing. Using LEMONADE, we detected somatic KRAS mutations in 129 of 179 (72%) cases, and showed that KRAS mutations were associated with bleeding as the first symptom (p=0.0072). Furthermore, we developed denovoSAVI (https://github.com/WangLabHKUST/denovoSAVI) to discover and prioritize de novo germline mutations, highlighting the critical role of ENG, EXPH5, JUP, and EPAS1 in bAVM development. To validate the role of de novo germline mutations, Qiuixia Zhou from Zilong's lab developed a zebrafish model to explore the role of de novo germline mutations in driving vascular malformation. Interestingly, knockdown of epas1 in zebrafish embryo showed AVM-like phenotype exclusively in the brain.
Moreover, leveraging bulk and single-cell sequencing data, we found abnormal expression of endothelial and mesenchymal markers in bAVM. This finding has been validated by flow-cytometric analysis and immunofluorescence staining, suggesting an involvement of Endothelial-to-Mesenchymal transition (EndMT) process in AVM. Following experimental studies demonstrated that somatic KRAS mutation and de novo germline mutations convergently promote endothelial-to-mesenchymal transition in this disease. Lastly, we showed that Lovastatin reversed EndMT features in vitro and ex vivo.
This study has made enormous contributions towards a clearer understanding of bAVM pathogenesis. The "anti-EndMT" treatment for AVM patients is therefore under further investigation.
Identification of MEKK3-I441M in cerebral cavernous malformation
Cerebral cavernous malformations (CCMs) are vascular disorders that affect up to 0.5% of the total population. About 20% of CCMs are inherited because of familial mutations in CCM genes, including CCM1/KRIT1, CCM2/MGC4607, and CCM3/PDCD10, whereas the etiology of a majority of simplex CCM-affected individuals remains unclear.
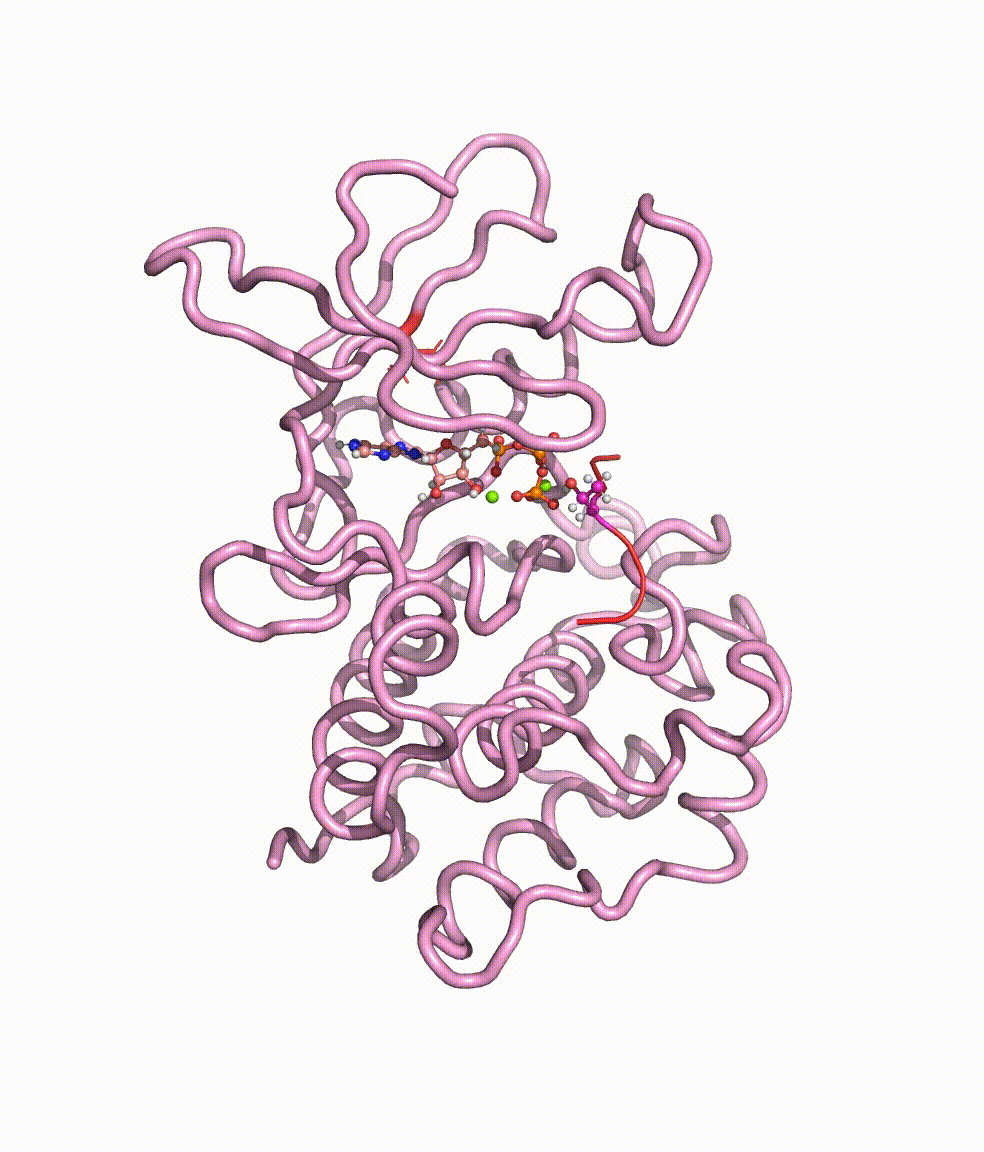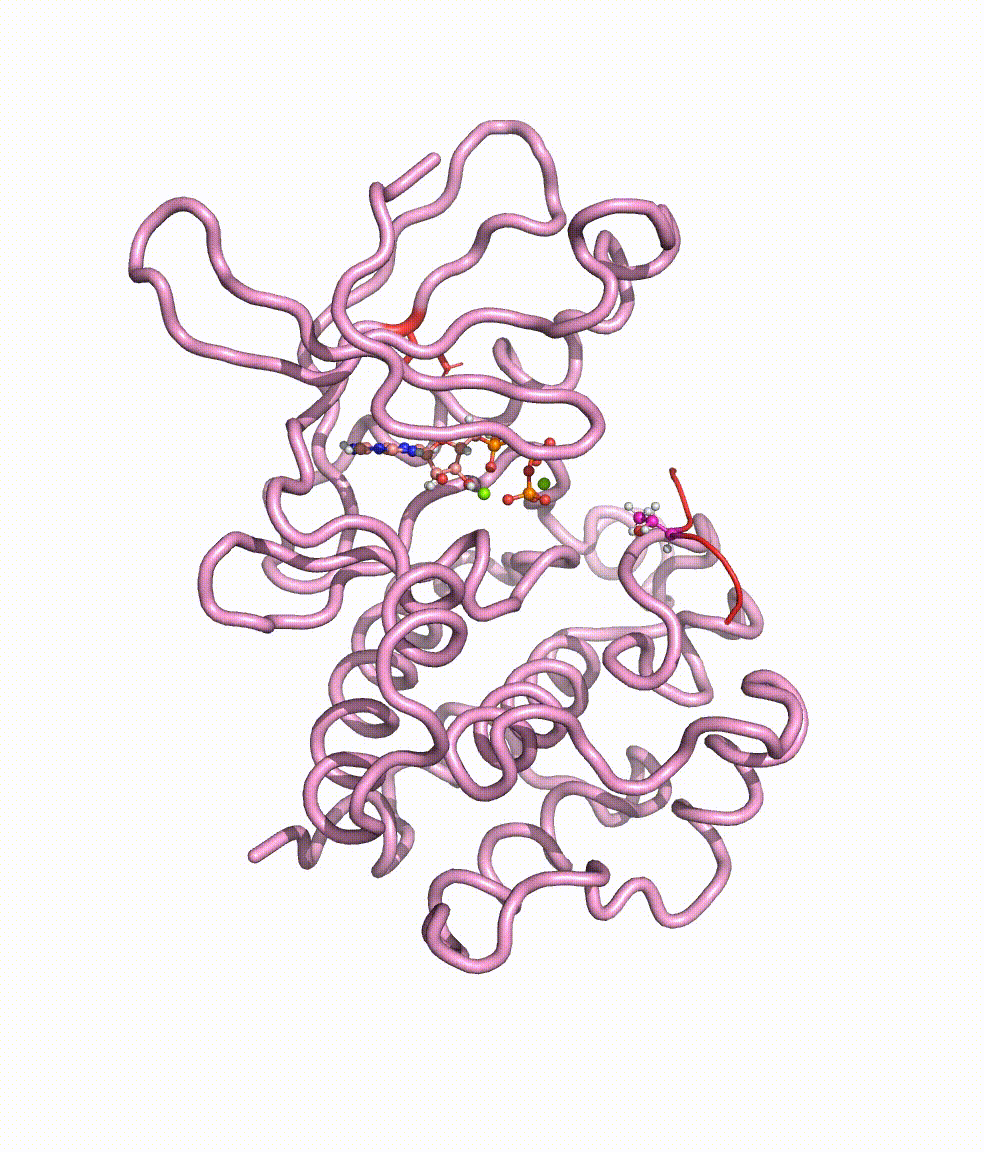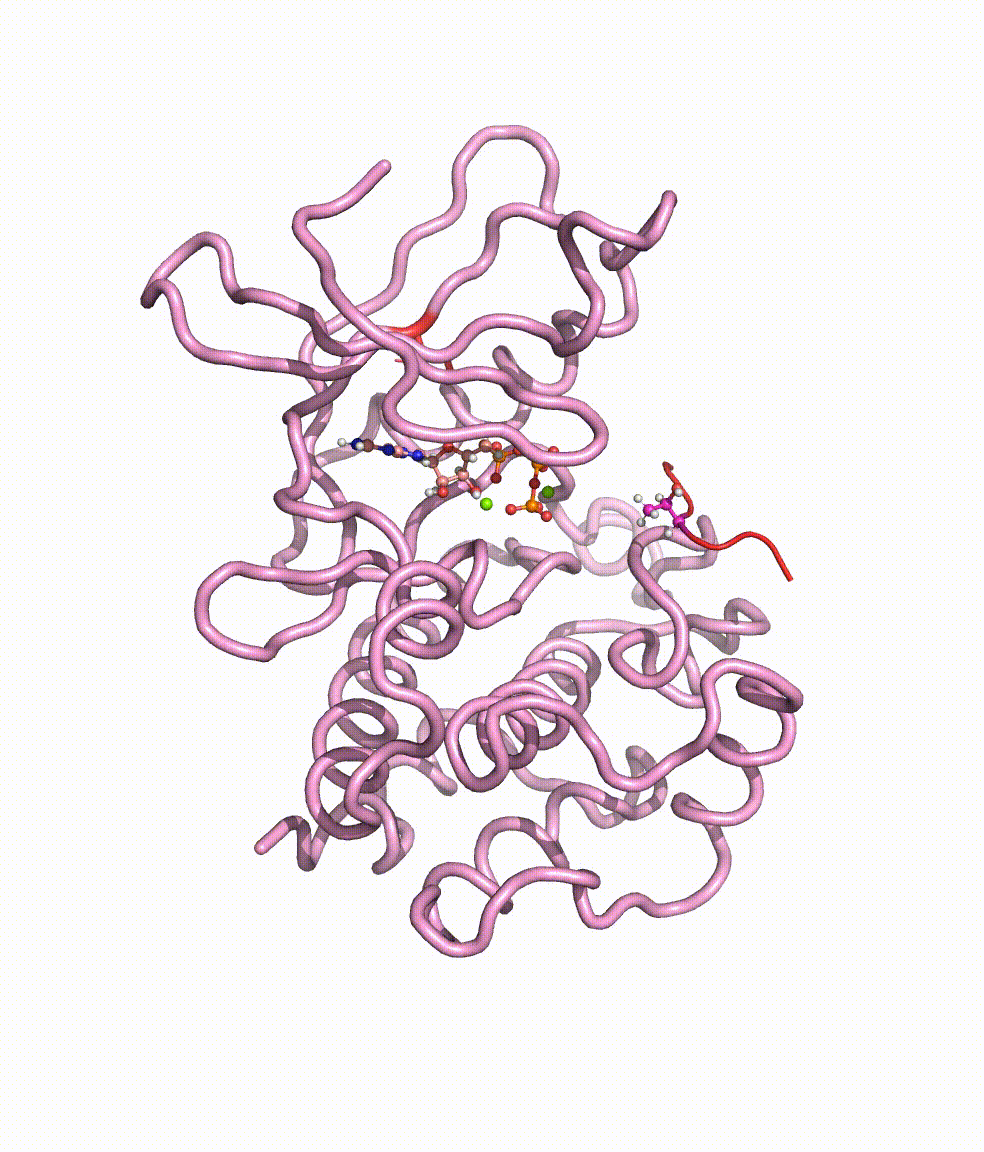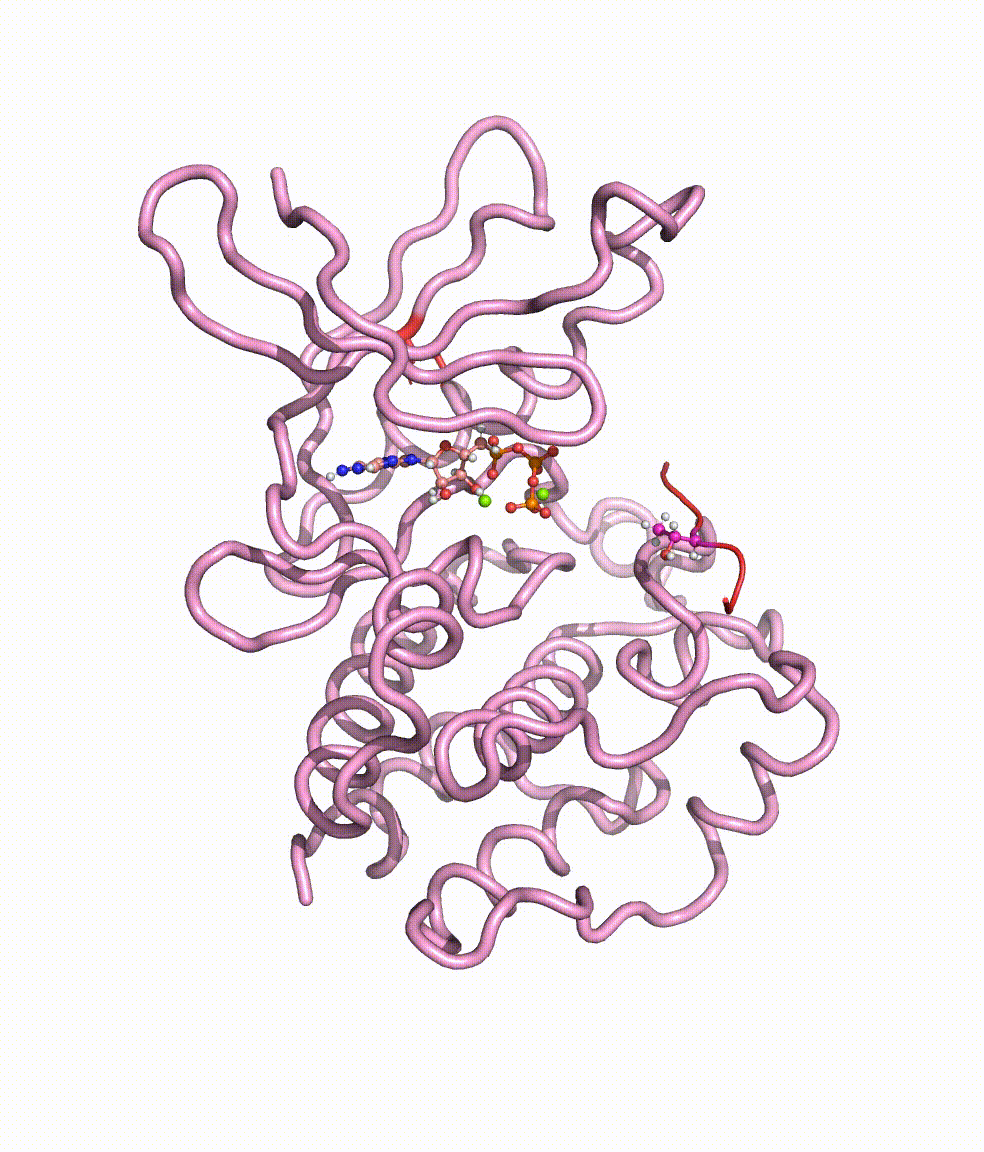
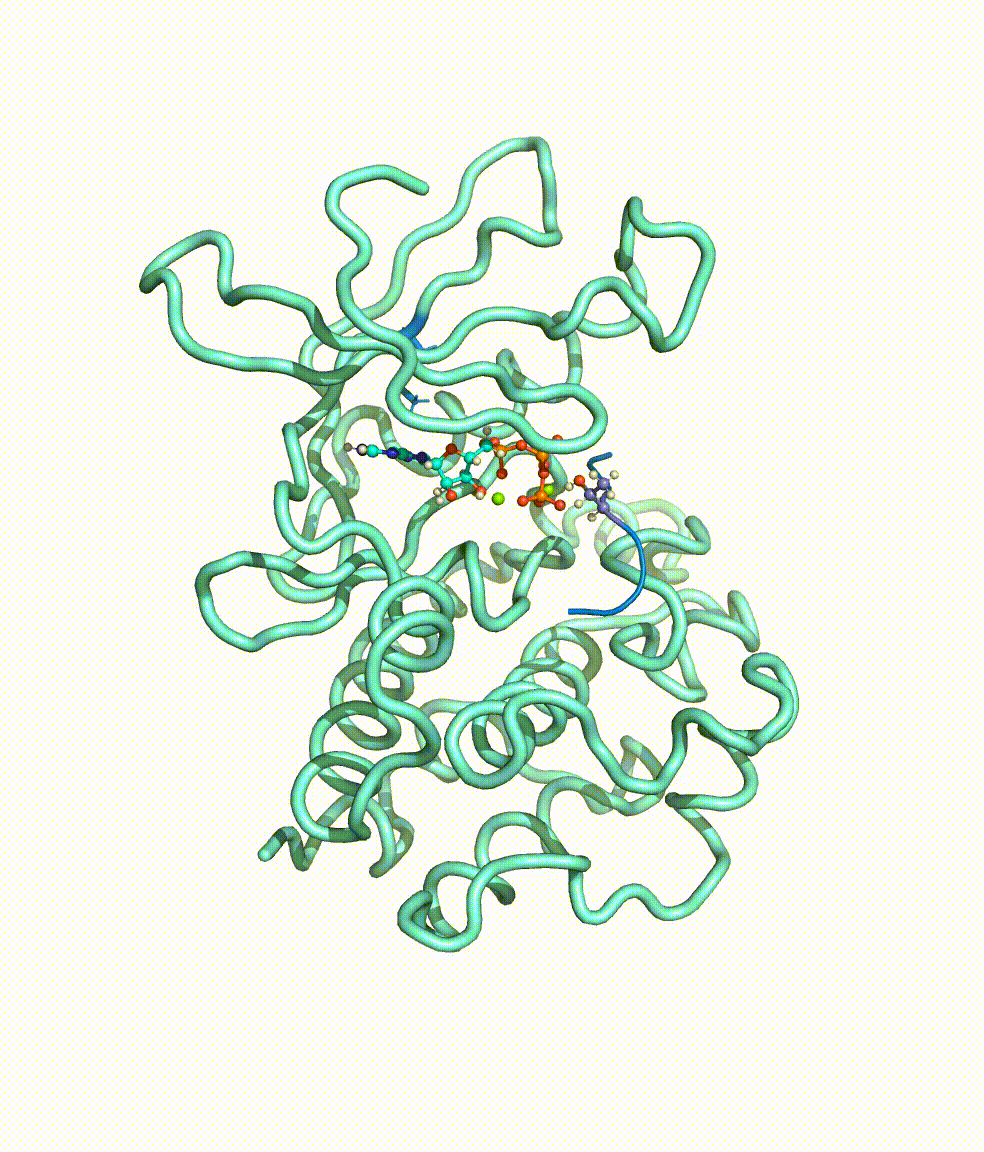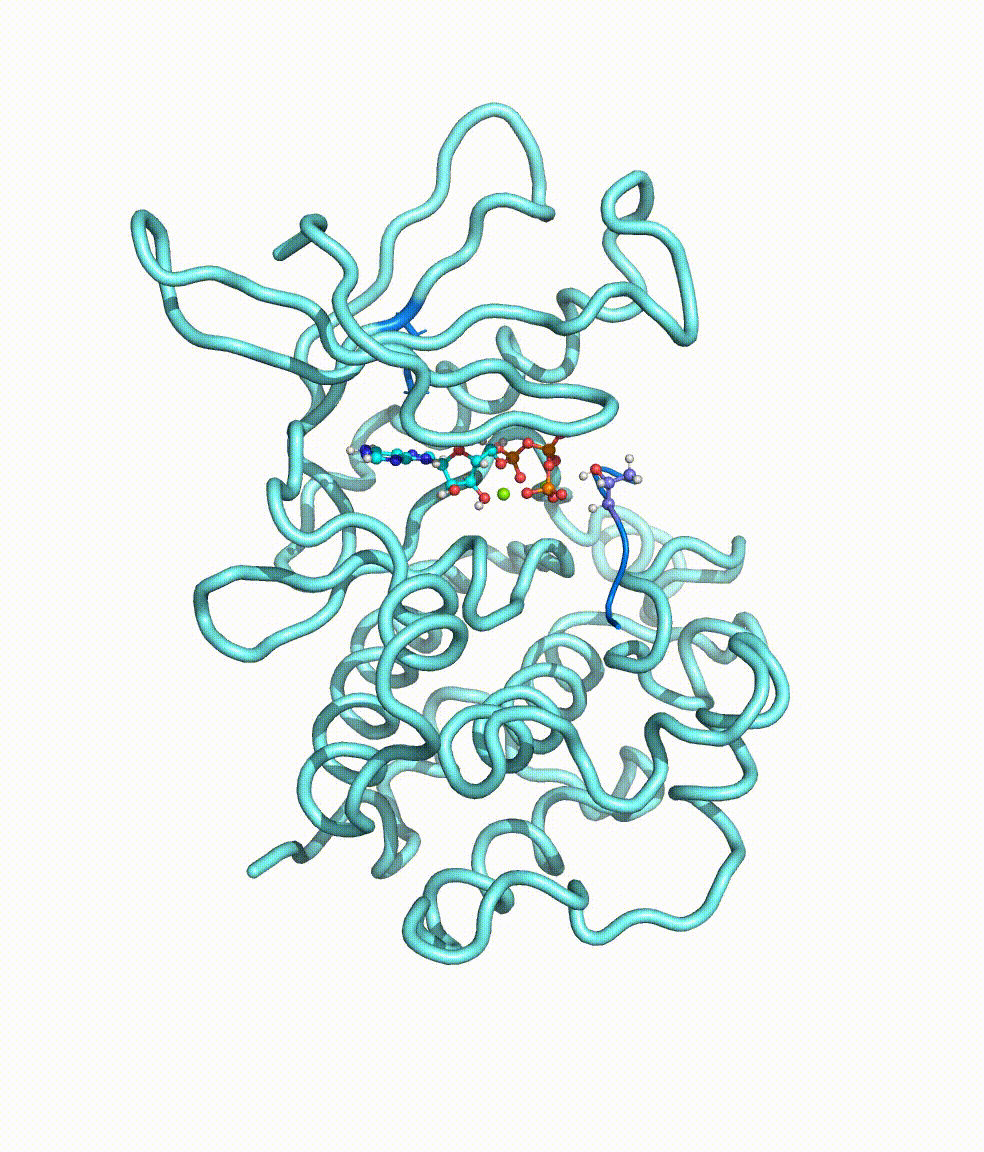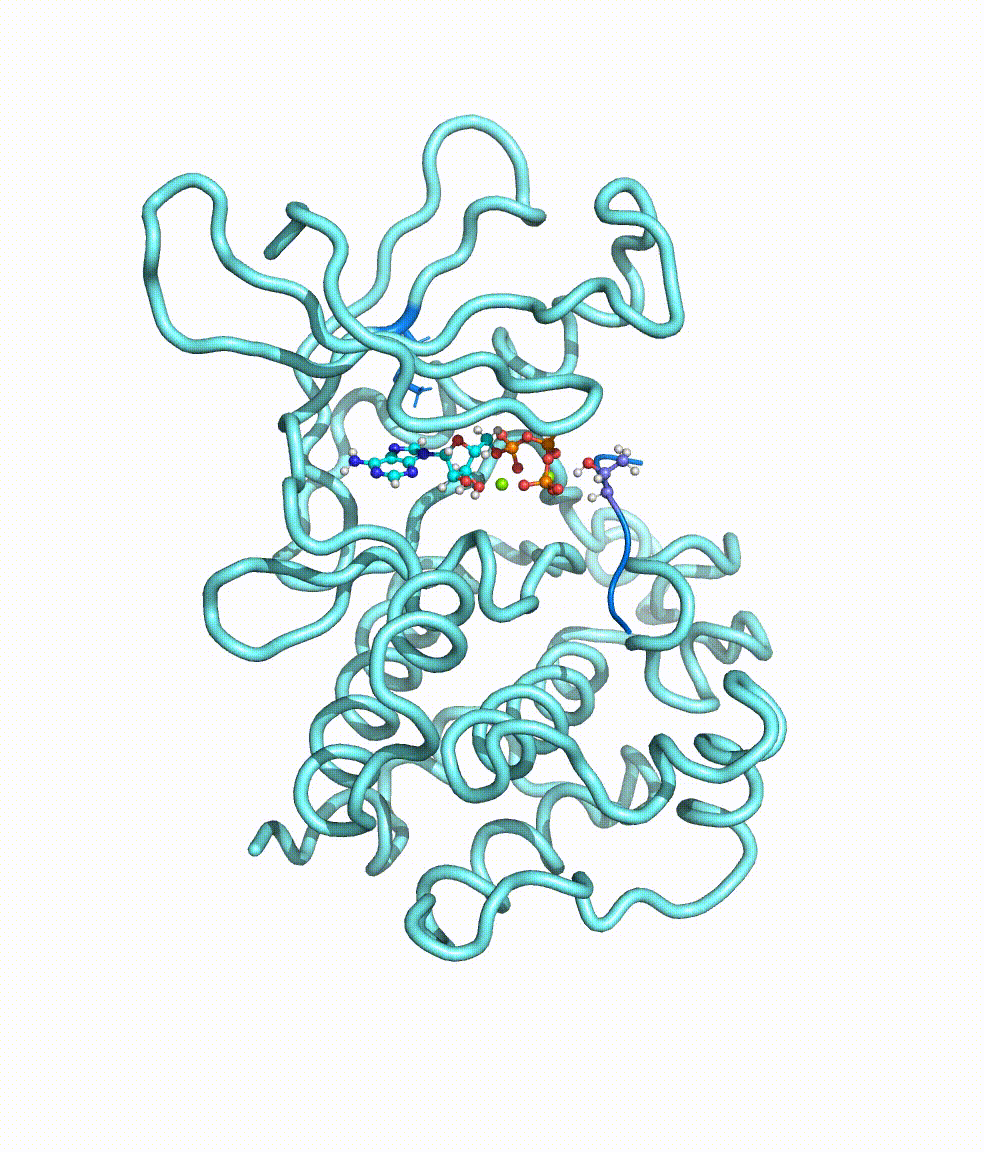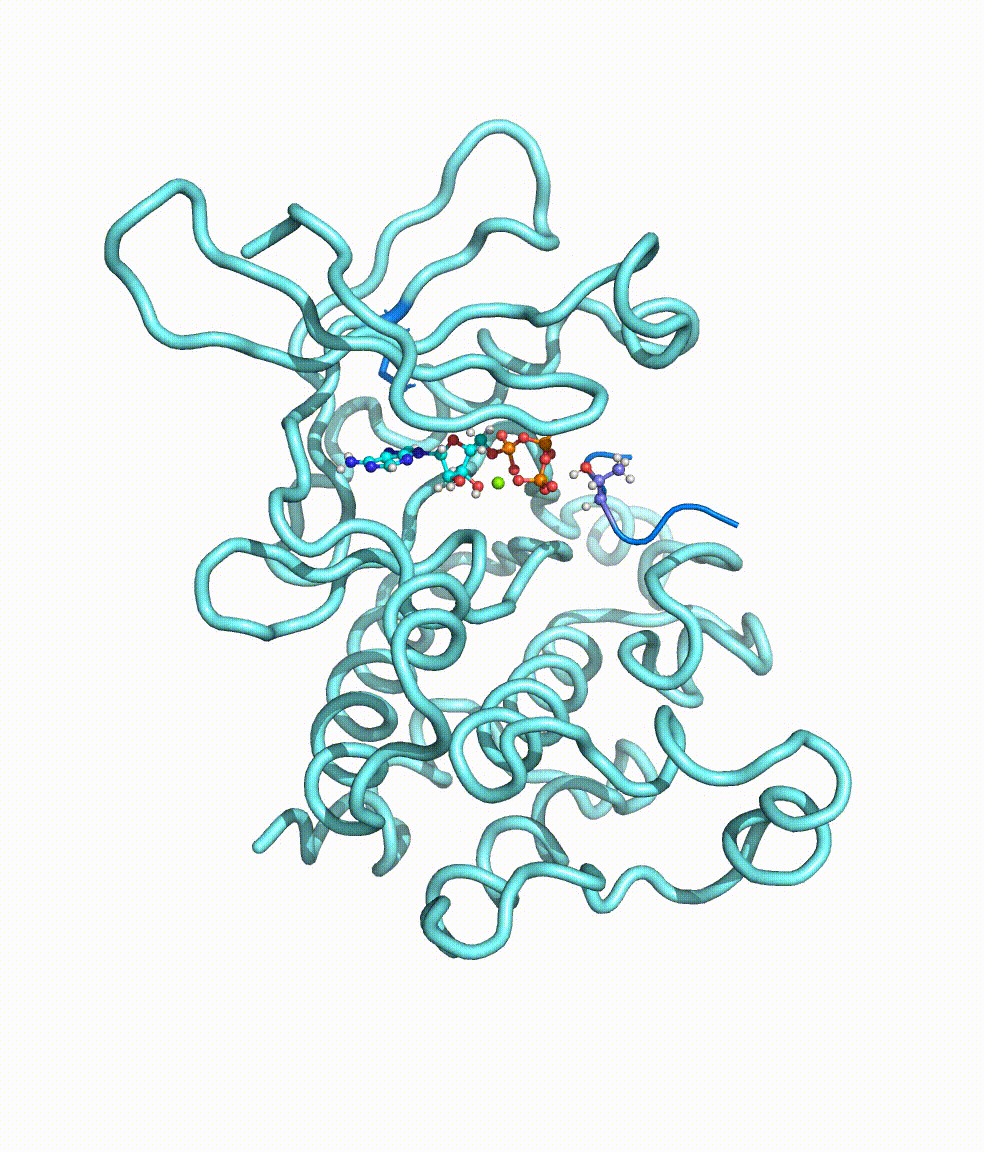
Collaborating with Dr. Yong Cao in Beijing Tiantan Hospital, we together revealed somatic mutations of MAP3K3, PIK3CA, MAP2K7, and CCM genes in CCM lesions. In particular, we discovered somatic hotspot mutations of PIK3CA in 11 of 38 individuals with CCMs, and MAP3K3 somatic mutation (c.1323C>G [p.Ile441Met]) in 37.0% (34 of 92) of the simplex CCM-affected individuals. Strikingly, we found that the MAP3K3 c.1323C>G mutation presents in 95.7% (22 of 23) of the popcorn-like lesions but only 2.5% (1 of 40) of the subacute-bleeding or multifocal lesions that are predominantly attributed to mutations in the CCM1/2/3 signaling complex. Leveraging mini-bulk sequencing, we demonstrate the enrichment of MAP3K3 c.1323C>G mutation in CCM endothelium. Notably, by using molecular-dynamics simulation, we demonstrated that the MAP3K3 c.1323C>G mutation driven MEKK3 (MAP3K3 encodes MEKK3) kinase domain conformation alteration enhanced the binding affinity of ATP, consequently leading to higher kinase activity in MEKK3 protein. Moreover, beyond the activation of CCM1/2/3-inhibited ERK5 signaling, MEKK3 p.Ile441Met also activates ERK1/2, JNK, and p38 pathways because of the mutation-induced MEKK3 kinase activity enhancement. Collectively, we identified several somatic activating mutations in CCM endothelium, and the MAP3K3 c.1323C>G mutation defines a primary CCM subtype with distinct characteristics in signaling activation and magnetic resonance imaging appearance. For more details please read the published paper in AJHG.
Watch vedio to know more on https://youtu.be/X5Re8-qjgQQ!

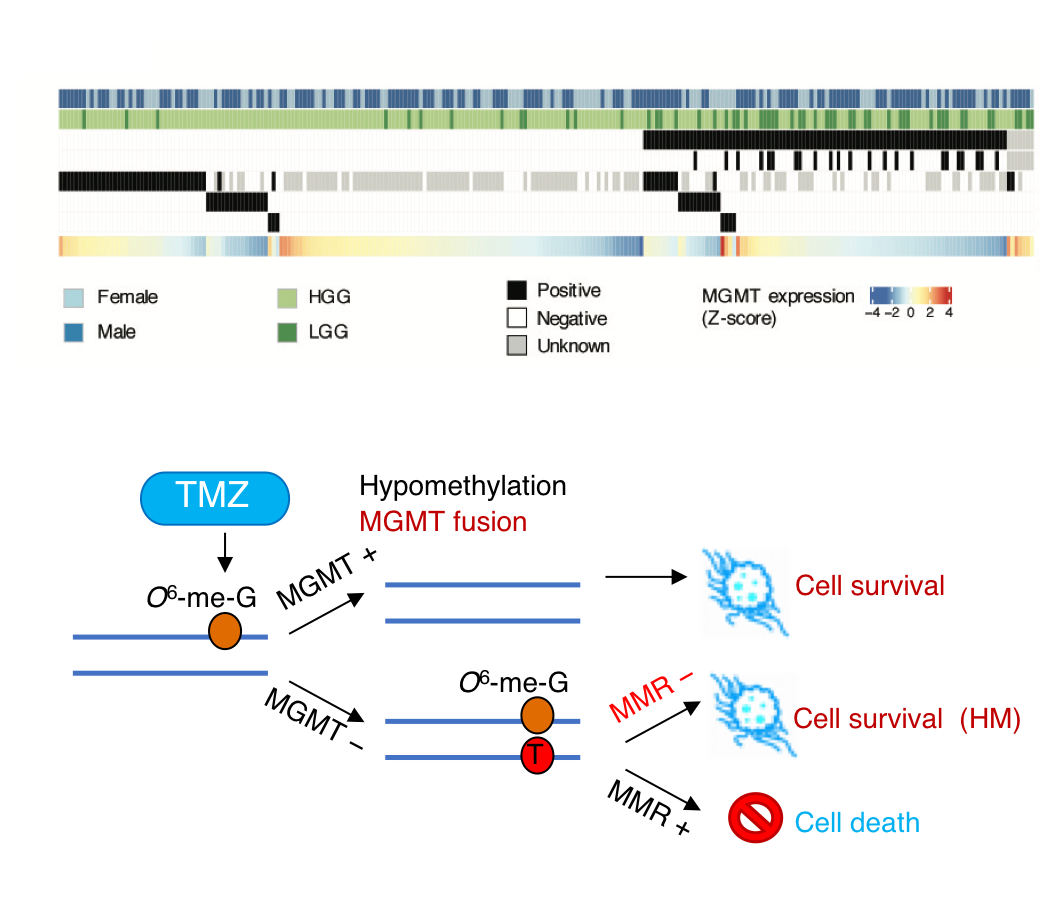
Genomic translocations of MGMT drive chemotherapy resistance in a
subgroup of glioma patients
Temozolomide (TMZ) is an oral alkylating agent widely used in the treatment of glioblastoma (GBM) and a group of high-risk low-grade glioma patients. The O-6-methylguanine-DNA methyltransferase (MGMT) is an intrinsic protein responsible for the direct repair of the main TMZ-induced toxic DNA damage. Here we collected and analysed RNA sequencing data of 252 recurrent gliomas with well-documented TMZ treatment history, and identified eight different MGMT gene fusions that are caused by genomic rearrangements, across all glioma subtypes. All fusions shared the same MGMT breakpoint that preserves the functional domains of this DNA methyltransferase. With more active promoters borrowed from gene-fusion partners, MGMT expression is significantly up-regulated, regardless of the methylation status of its original promoter. Interestingly, we also observed that the detected MGMT fusion, MGMT promoter hypomethylation, and therapy-associated somatic hypermutation tend to be mutually exclusive, suggesting that these events might be alternative mechanisms leading to TMZ resistance. In vitro and in vivo models generated by Squatrito group provided solid evidence of the role of MGMT genomic rearrangements in TMZ resistance. For more details please read the published paper in Nature Communications.
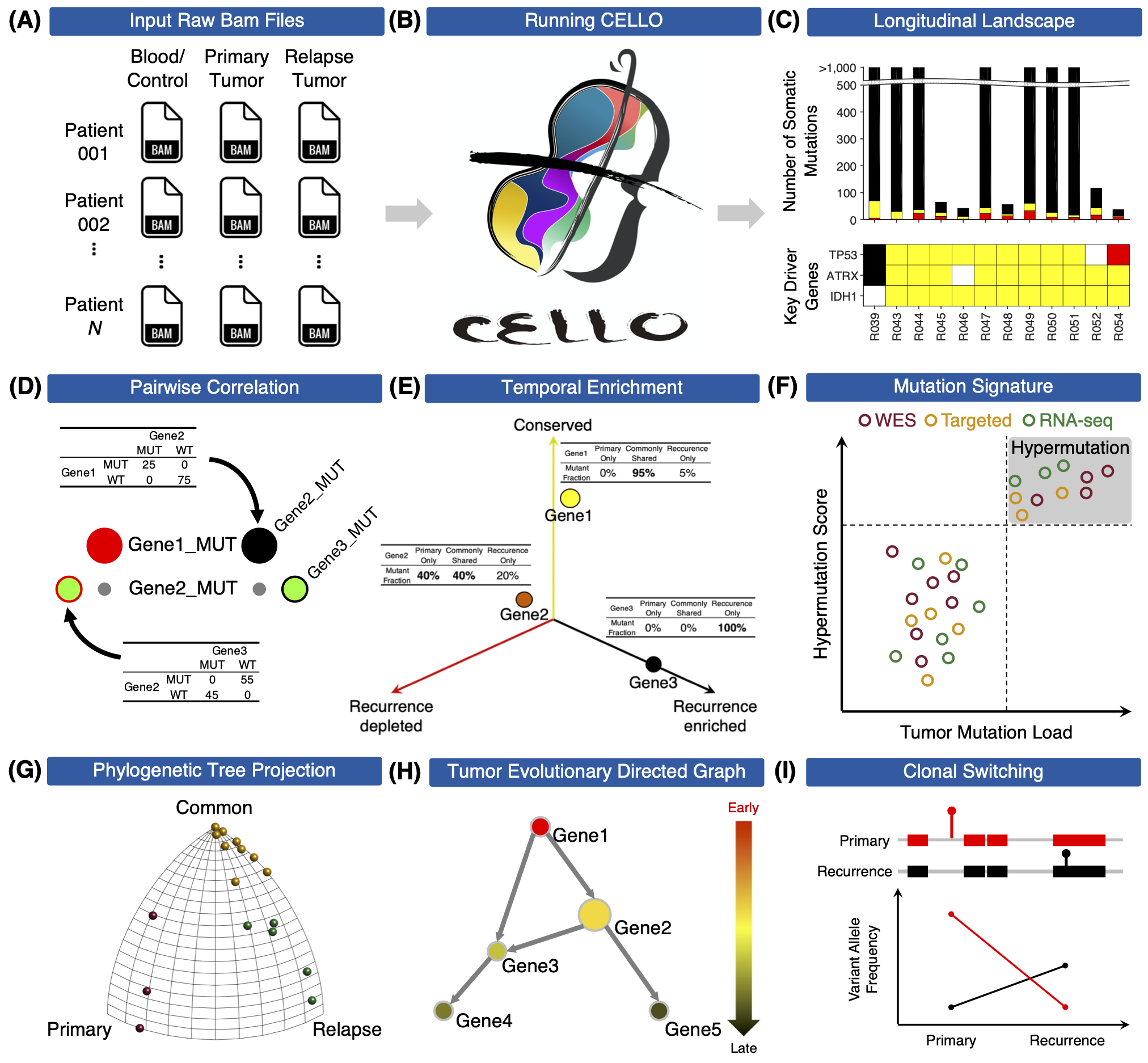
Tools for untangling cancer evolution from longitudinal genomic data
Targeting tumor-specific mutations via customized chemical compounds can precisely eradicate the cancer cells without harming healthy tissues, which paves a way toward precision oncology. But this precision oncology strategy has not been successful in many refractory cancers such as glioblastoma (GBM). One of the main obstacles is the limited understanding of cancer evolution, in which cancer cells might acquire advantageous fitness to revive under treatment stress.
To understand how cancer evolves under treatment stress, the Wang Lab developed CELLO (Cancer EvoLution for LOngitudinal data), to analyze and visualize longitudinal next-generation sequencing data before and after treatment. Particularly, CELLO can conduct the following analytical workflow including (1) generation of longitudinal mutational landscape, (2) detection of mutational signature for cross-platform sequencing data, (3) clustering of patients based on evolutionary patterns, (4) identification of clonal switching events; and (5) inference temporal order of somatic mutations. To benefit researchers who are interested in longitudinal cancer genomics study for analyzing their own data, both MATLAB and R versions of CELLO are developed. To ensure reproducibility and usability, we also present a docker version of CELLO based on the R implementation. For more details please read the published paper in Quantitative Biology.
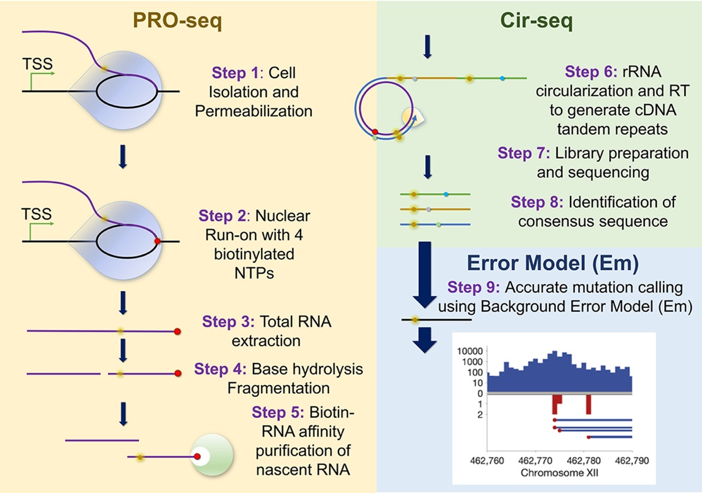
New sequencing protocol and model identify transcription errors
RNA polymerase transcribes certain genomic loci with higher errors rates. These transcription error-enriched genomic loci (TEELs) have implications in disease. Current deep-sequencing methods cannot distinguish TEELs from post-transcriptional modifications, stochastic transcription errors, and technical noise, impeding efforts to elucidate the mechanisms linking TEELs to disease.
Collaborating with Prof Xuhui Huang in HKUST Chemistry, we together describe background error model-coupled precision nuclear run-on circular-sequencing (EmPC-seq) to discern genomic regions enriched for transcription misincorporations. Applying EmPC-seq to the ribosomal RNA transcriptome, we show that TEELs of RNA polymerase I are not randomly distributed but clustered together, with higher error frequencies at nascent transcript 3′ ends. Our study establishes a reliable method of identifying TEELs with nucleotide precision, which can help elucidate their molecular origins. For more details please read the published paper in Journal of Molecular Biology.
Classifying gastric cancer using FLORA reveals clinically-relevant
molecular subtypes and highlights LINC01614 as a biomarker for
patient prognosis
In recent years, long non-coding RNAs (lncRNAs) have been extensively studied, revealing their diverse functions in tumor progression, metastasis and drug resistance. Through ab initio assembly, it is possible to find previously unknown lncRNAs with important functions. LncRNA analysis pipelines including ATRAIN (Cell 2015) and NORI (Gut 2019) have incorporated the transcript assembly step into their workflows. The FLORA pipeline developed by WangLab is a user-friendly computational pipeline that accelerates the process of lncRNA discovery as well as provides functional predictions for lncRNAs, which would be useful for lncRNA analysis in large cohorts of cancer patients and prioritization of potential oncogenic lncRNAs.
Applications of the FLORA pipeline on 375 gastric cancer (GC) and 27 tumor-adjacent samples in the TCGA cohort, our study revealed the comprehensive landscape of lncRNAs in GC and revealed 1,547 novel lncRNAs that has not been discovered or annotated. Clustering GC patients based on 1,235 tumor-specific lncRNAs further revealed three tumor subtypes, and the lncRNA-based subtype 3 (L3 subtype) was associated with poor survival across multiple cohorts of patients. The lncRNA-based subtype is an independent prognosis factor in GC and reveal a subset of intestinal-histology patients with worse survival. Moreover, tumor-specific lncRNAs also have great potentials to be developed into GC biomarkers. Specifically, LINC01614 is highlighted as the top candidate, as LINC01614 expression is highly tumor-specific and strongly associated with poor prognosis and tumor metastasis. We further verified the functions of LINC01614 in promoting GC proliferation and metastasis through over-expression and CRISPR-Cas9 knockout experiments, and revealed potential downstream targets of LINC01614. For more details, please read the published paper in Oncogene.







